Yeast Display - FACS Sorting
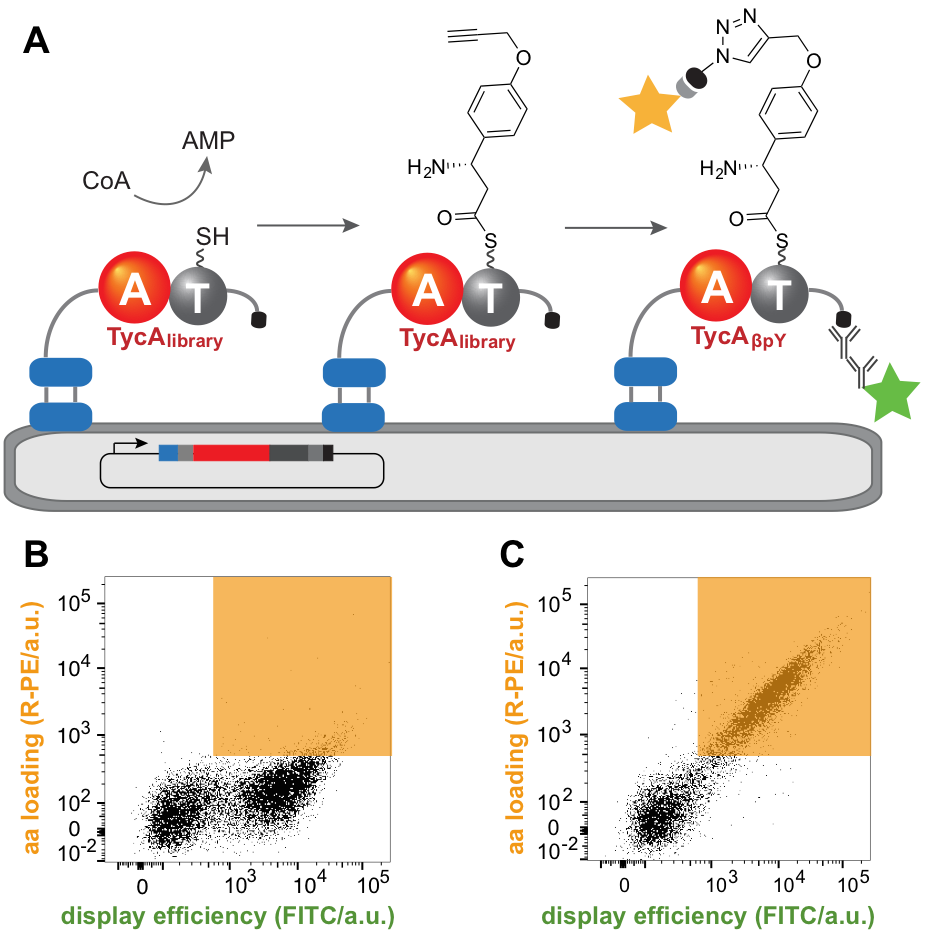
The adenylation (A) domains of NRPS function as gatekeepers, selectively loading individual amino acid building blocks onto the pantetheine arm of the enzyme. Thus, their specificity dictates the final peptide sequence and engineering their substrate preferences is the first step towards tailored peptides. Substrate loading entails transiently attaching the substrate covalently to the enzyme, so displaying the enzyme on yeast cells and using taggable substrates provides a simple mean to link genotype and phenotype. To that end, we developed an adenylation domain that activates an unnatural, “clickable” amino acid that can be derivatized with a fluorophore to provide a convenient readout. Active variants could be isolated from libraries containing up to 108 members by fluorescence-activated cell sorting (FACS). The ability to introduce multiple mutations simultaneously facilitates identification of beneficial synergistic interactions and supersedes numerous rounds of evolution using lower throughput methods. This approach was successfully used to reprogram the Phe-activating A domain of an NRPS system to introduce (S)-β-phenylalanine with a 40,000-fold switch in α/β specificity, highlighting its great potential.
Selected Publications
Kries, H. Biosynthetic engineering of nonribosomal peptide synthetases. Journal of Peptide Science 22, 564-570 (2016).
Niquille, D. L. et al. Nonribosomal biosynthesis of backbone-modified peptides. Nature Chemistry 10, 282-287 (2018).